Mapache
Mapache ([maˈpa.t͡ʃe]) is a lightweight mapping pipeline for ancient DNA using the workflow manager Snakemake .
Visit the Wiki for extensive documentation on how to use mapache and follow the tutorial to map and impute the test dataset.
If you already have some experience with DNA mapping and/or Snakemake, you can follow the quick guide below.
The usage of this workflow is described in the Snakemake Workflow Catalog .
The following main steps are included in mapache :
for each fastq file:
- subsampling of a small subset of reads (options, not run by default; useful for quick tests)
- adapter removal with AdapterRemoval (optional; run by default)
- mapping with bwa aln (default), bwa mem or bowtie2
for each library:
- marking duplicates with MarkDuplicates (optional; duplicates are removed by default)
- rescaling of base qualitiy scores with mapDamage (optional, not run by default)
for each sample:
- re-alignment of reads with GATK (optional; run by default)
- re-computation of the md flag with samtools (optional; run by default)
- imputation of low-coverage genomes with GLIMPSE (optional, not run by default)
Moreover, mapache allows you to map large datasets to a single reference genome or to multiple genomes.
Mapache is designed having in mind that one or multiple DNA libraries are generated per sample, and such libraries are sequenced at least once (e.g., for screening), but usually multiple times (normally prioritizing the highest-quality libraries). This results in several FASTQ files, which have to be mapped several times over the course of a project in order to generate a single BAM file.
The goal of mapache is to make the mapping process as easy and transparent as possible, whether you are mapping to a single or multiple genomes, mapping for the first time, needing to update a BAM file or even impute low-coverage genomes.

Quick guide
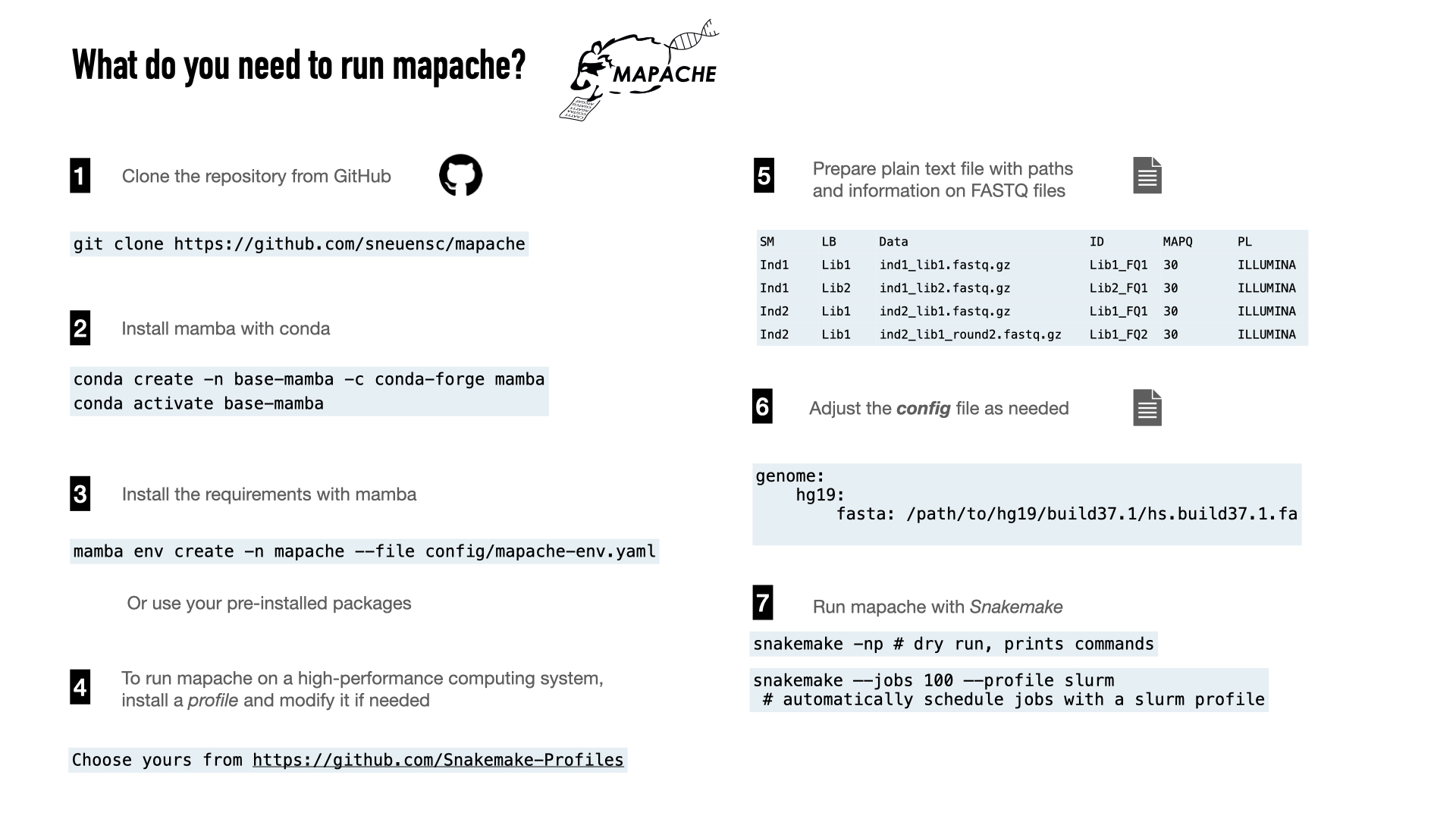
To install mapache, you need to clone the repository and create a conda environment following the instructions below. This will automatically install the software needed to map FASTQ files to a reference genome and create a report with the mapping statistics.
To execute mapache, you can move to the cloned directory (
mapache
) or symlink its content (the directories
config/
,
results/
,
workflow/
, and
test_data/
if you want to run the test) to your working directory.
You will mostly interact with mapache through its
configuration file
(
config/config.yaml
), where you can tweak the parameteres of the pipeline, and the
samples file
(
config/samples.tsv
), in which you can list all the input FASTQ files.
Installation
#--------------------------------------------------------------#
## create mamba environment
conda create -n base-mamba -c conda-forge mamba
## activate mamba environment
conda activate base-mamba
#--------------------------------------------------------------#
## clone mapache repository
git clone https://github.com/sneuensc/mapache.git
cd mapache
## create conda environment for mapache
mamba env create -n mapache --file config/mapache-env.yaml
#--------------------------------------------------------------#
## now you can activate the mapache environment
conda activate mapache

Prepare samples file
A samples file for the test dataset is provided (
config/samples.tsv
). If you want to run it on your own datasets, you can prepare a tab-separated file containing the same columns as the example.
Example of a sample file for single-end libraries:
SM LB ID Data
ind1 L1 L1_1 reads/ind1.L1_R1_001.fastq.gz
ind1 L1 L1_2 reads/ind1.L1_R1_002.fastq.gz
ind1 L2 L2_1 reads/ind1.L2_R1_001.fastq.gz
ind1 L2 L2_2 reads/ind1.L2_R1_002.fastq.gz
Example of a sample file for paired-end and single-end libraries:
SM LB ID Data1 Data2
ind2 L1 L1_1 reads/ind2.L1_R1_001.fastq.gz reads/ind2.L1_R2_001.fastq.gz
ind2 L1 L1_2 reads/ind2.L1_R1_002.fastq.gz reads/ind2.L1_R2_001.fastq.gz
ind2 L2 L2_1 reads/ind2.L2_R1_002.fastq.gz NULL
In the first example, four fastq files will be mapped. They were generated from two different libraries (here, labelled as
L1
and
L2
) from a single sample (
ind1
).
In the second example, there is still only one sample (
ind2
), and two libraries, sequenced in paired-end (
L1
) and single-end (
L2
) mode.
You can add more samples/libraries/fastq files in the input dataset by adding a new row including the 4 or 5 fields indicated above.
The columns of the sample file are:
-
SM: Sample name. Libraries are merged according to this name.
-
LB: Library name. Fastq files are merged according to this name.
-
ID: An ID for the fastq library (examples: id1, fq_1, ind1_lib1_fq2, etc.)
-
Data (single-end format) : Path to the fastq file. The file may be gzipped or not. Path may be absolute or relative to the working directory.
-
Data1 (paired-end format) : Path to the forward fastq file (R1) for paired-end data or the fastq file for single-end data. The file may be gzipped or not. Path may be absolute or relative to the working directory.
-
Data2 (paired-end format) : Path to the reverse fastq file (R2) for paired-end data or
NULLfor single-end data. The file may be gzipped or not. Path may be absolute or relative to the working directory.
👆🏿 Note that you can select any label you want for the columns SM, LB, and ID, but they may not contain points '.'. That is, they do not need to match any substring or the name of your FASTQ files (FASTQ file names may in contrast contain points). However, we recommend that you stick to meaningful names (e.g.: Denisova, Mota, UstIshim, Anzick, lib1, lib2, lib3_USER, etc.) and ACII characters.
Adapt config file
By default, mapache is configured to map ancient data to a human reference genome. You need to specify the path to the reference genome in FASTA format in the configuration file (
config/config.yaml
) provided by mapache.
Specify reference genome
Mapache will map the input FASTQ files to the reference genome(s) indicated in the config file under the keyword
genome
.
The path to the genome in FASTA format should be indicated with the keyword
fasta
.
In the example below, all the samples will be mapped to two versions of the human genome. The final BAM files will be named with the format
{sample_ID}.{genome_name}.bam
(e.g., ind1.hg19.bam, ind1.GRCh38.bam).
genome:
GRCh37: path_to_reference/GRCh37.fasta
# GRCh38: path_to_reference/GCA_000001405.15_GRCh38.fa
Enable/disable steps and specify memory and runtime
Mapache will perform different steps (see below) in order to produce a BAM file. Most of the steps are optional, but run by default.
The config file contains entries corresponding to each of the steps that can be run and customized. For example, the entry with the options to clean the fastq files looks like this:
# fastq cleaning (optional)
cleaning:
run: 'adapterremoval' # options: adapterremoval (default), fastp, False
params_adapterremoval: '--minlength 30 --trimns --trimqualities'
params_fastp: ''
threads: 4
mem: 4 ## in GB
time: 2
If you need to modify the pipeline, for instance to ommit the step step, you can set in the config file its option to
run: False
, or
run: 'adapterremovel'
and
run: 'fastp'
to run
AdapterRemoval2
and
fastp
, respectively. Additionally, if you intend to run mapache on an HPC system, you can specify the number of
threads
, memory (
mem
in GB), and runtime (
time
in hours) to be allocated to each step. Finally, you can pass additional parameters to the tool to be executed (e.g.
--minlength 30
to AdapterRemoval2) with the keyword
params_adapterremoval
.
Running mapache
To run mapache on your working directory, you will need to copy or create symbolic links to the following directories:
-
workflow (mandatory)
-
config (mandatory)
-
test_data (optional, only if you want to run the tutorial)
-
slurm (optional, useful to submit jobs in HPC systems with slurm as cluster manager)
Assuming that you are in the mapache directory, you can use the next lines to create an alias to copy or symlink those directories. You can adjust it by including only the directories that you want to copy/symlink.
# alias to cp directories
echo "alias copy_mapache='cp -r $(pwd -P)/{config,workflow,test_data,slurm} . ' " >> ~/.bash_profile
# alias to symlink directories
echo "alias symlink_mapache='ln -s $(pwd -P)/{config,workflow,test_data,slurm} . ' ">> ~/.bash_profile
source ~/.bash_profile

Make sure that the paths to
-
the reference genome(s) in the config files
-
the file listing the FASTQ files (samples.tsv)
as well as the paths listed in samples.tsv are properly specified
Dry run
We highly recommend to make use of dry runs to get an idea of the jobs that will be executed.
# print jobs to be executed
snakemake -pn
# Visualization of the pipeline in a directed acyclic graph (DAG).
snakemake dag --dag | dot -Tpng > dag.png
The command below will produce a figure with the rules that will be run.
snakemake --rulegraph |dot -Tpng > rulegraph.png

Examples for the DAG and rulegraph can be found here and here , respectively.
Optional: configure a profile to automatically submit jobs to a queue
This is a very handy utility that will allow mapache to automatically schedule jobs for submission to a queuing system. If you work with
a system managed by slurm
, you can
skip this step
, as mapache comes with a slurm profile (
mapache/slurm
).
Other profiles are available at: https://github.com/Snakemake-Profiles
Note however that the exact configuration might vary depending on your system (i.e., some users might have different accounts on a server for billing purposes). We thus highly encourage you to seek for help with your IT team if you are in doubt about the configuration that suits your system the best. Please note that we are not in charge of developing these profiles.
Run mapping pipeline
mapache can be run locally by indicating the number of cores available or with a high-performance computing system, by configuring a profile.
Example of execution using only one CPU:
snakemake --cores 1

If you work on an HPC system managed by slurm, you can use the slurm profile in the repository (
mapache/slurm/
) by symlinking it to your working directory. We recommend to start a screen session prior to the job submission.
Example of a submission of 999 jobs (simoultaneously) with the slurm profile:
snakemake --jobs 999 --profile slurm
Create report with mapping statistics
The template using to produce the html report will change depending on the version of Snakemake that you are using with
mapache
. Although we try to include one of the most recent distributions of Snakemake, we think that the template used in older versions is more useful to navigate through the results in the report. We thus recommend you to create a new environment with conda or mamba to create such report:
mamba create -n snakemake610 snakemake=6.10.0
mamba activate snakemake610
Once you have activated the new environment, creating the report is straightforward:
snakemake --report report.zip
or
snakemake --report report.html
We recommend creating the zip version of the report, as it contains the html report in it and it allows you to download any of the output tables or plots by clicking on the links of the report, making it easier to share with your colleagues.
Explore the zipped report produced with Snakemake 6.10.0.
Summary of useful commands
#------------------------------------------
# Get an idea of the jobs that will be executed. Not mandatory but very useful to spot possible mistakes in the configuration or input files
snakemake dag --dag | dot -Tsvg > dag.svg Visualization of the pipeline in a directed acyclic graph (DAG).
snakemake dag --rulegraph | dot -Tsvg > rulegraph.svg Visualization the interplay of the rules.
snakemake -n Dry run
snakemake -p -n Print out the commands in a dry run
#------------------------------------------
# recommended workflow on a cluster (e.g. slurm)
snakemake --jobs 999 --profile slurm Run all mappings and imputation (note that the profile has to be the last argument)
#------------------------------------------
# recommended workflow on a single machine without a queuing system
snakemake --cores 16 Run all mappings and imputation (assuming that you have 16 CPUs available)
#------------------------------------------
# Create report (after execution)
snakemake --report report.html Create a html report
snakemake --report report.zip Create a zip report; useful if you want to download the files by clicking on the HTML report
#------------------------------------------
# Options to control the execution.
# Visit https://snakemake.readthedocs.io/en/stable/ for full documentation
--profile slurm To send it on the cluster (must be the last argument).
-R RULE_NAME Force a start at least at the given rule.
--until RULE_NAME Run until the given rule (included).
--rerun-incomplete Re-run all jobs the output of which is recognized as incomplete.
--configfile FILE Define the config file (default config/config.yaml).
-t Reset the timestamp that the output is not re-computed.
-p Print out the shell commands that will be executed.
--rerun-triggers mtime for a rerun, consider only the time stamp (previously the default
setting, now any change (code, config,..) lead to a rerun of
the rule)
Citing mapache
If you use mapache in your study, please cite Neuenschwander S, Cruz Dávalos DI, et al. (2023) Mapache: a flexible pipeline to map ancient DNA. Bioinformatics (https://doi.org/10.1093/bioinformatics/btad028) and the tools that you used within mapache. See the table below for a list of tools used at each step.
Tools included in mapache
| Version | Reference | Link | |
|---|---|---|---|
| Workflow manager | |||
| Snakemake | 7.18.2 | Mölder, et al. (2021) | https://github.com/snakemake/snakemake |
| Subsample | |||
| seqtk | 1.3 | https://github.com/lh3/seqtk | |
| Clean | |||
| AdapterRemoval2 | 2.3.2 | Schubert, et al. (2016) | https://github.com/MikkelSchubert/adapterremoval |
| fastp | 0.23.2 | Chen, et al. (2018) | https://github.com/OpenGene/fastp |
| Map | |||
| BWA aln | 0.7.17 | Li and Durbin (2009) | https://github.com/lh3/bwa |
| BWA mem | 0.7.17 | Li and Durbin (2009) | https://github.com/lh3/bwa |
| Bowtie2 | 2.4.4 | Langmead and Salzberg (2012) | https://github.com/BenLangmead/bowtie2 |
| Sort | |||
| SAMtools | 1.14 | Danecek, et al. (2021) | https://github.com/samtools/samtools |
| Filter | |||
| SAMtools | 1.14 | Danecek, et al. (2021) | https://github.com/samtools/samtools |
| Merge lanes | |||
| SAMtools | 1.14 | Danecek, et al. (2021) | https://github.com/samtools/samtools |
| Remove duplicates | |||
| Picard MarkDuplicates | 2.25.5 | Broad Institute (2019) | http://broadinstitute.github.io/picard |
| dedup | 0.12.8 | Peltzer, et al.(2016) | https://github.com/apeltzer/DeDup |
| Rescale damage | |||
| mapDamage2 | 2.2.1 | Jonsson, et al. (2013) | https://github.com/ginolhac/mapDamage |
| Trim alignments | |||
| BamUtil | 1.0.15 | Jun, et al. (2015) | https://github.com/statgen/bamUtil |
| Merge libraries | |||
| SAMtools | 1.14 | Danecek, et al. (2021) | https://github.com/samtools/samtools |
| Realign indels | |||
| GATK IndelRealigner | 3.8 | DePristo, et al. (2011) | https://gatk.broadinstitute.org |
| Recompute md flag | |||
| SAMtools | 1.14 | Danecek, et al. (2021) | https://github.com/samtools/samtools |
| Imputation | |||
| GLIMPSE | 1.1.1 | Rubinacci, et al. (2021) | https://github.com/odelaneau/GLIMPSE |
| BCFtools | 1.15 | Danecek, et al. (2021) | https://github.com/samtools/bcftools |
| Reports | |||
| FastQC | 0.11.9 | Andrews (2010) | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| Qualimap | 2.2.2d | Okonechnikov, et al. (2016) | http://qualimap.conesalab.org |
| MultiQC | 1.13 | Ewels, et al. (2016) | https://multiqc.info |
| Statistics | |||
| BEDTools | 2.30.0 | Quinlan and Hall (2010) | https://github.com/arq5x/bedtools2 |
| bamdamage | modified | Malaspinas, et al. (2014) | https://savannah.nongnu.org/projects/bammds |
| R | 4.0 | R Core Team (2022) | https://www.r-project.org |
Code Snippets
30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 | shell: """ ## download file wget -O {output} {params.ftp} > {log}; ## test md5sum if available if [ "{params.md5}" == "''" ] || [ "{params.md5}" == "nan" ] ; then echo "WARNING: Downloaded fastq file '{params.ftp}' has no md5sum to verify the download!"; else if [ $(md5sum {output} | cut -d' ' -f1) != "{params.md5}" ]; then echo "ERROR: Downloaded fastq file '{params.ftp}' has a wrong md5sum!"; exit 1; fi fi """ |
74 75 | script: "../scripts/subsample_fastq.py" |
92 93 94 95 | shell: """ ln -srf {input} {output} """ |
154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 | shell: """ out={output.R}; AdapterRemoval --threads {threads} {params.params} --file1 {input[0]} \ --file2 {input[1]} --basename ${{out%%.fastq.gz}} --gzip \ --output1 {output.R1} --output2 {output.R2} \ --outputcollapsed {output.col_R} \ --outputcollapsedtruncated {output.col_trunc} 2> {log}; ## what should be retained options={params.collapsed}; if [[ "$options" == "only_collapse" ]]; then ln -srf {output.col_R} {output.R}; elif [[ "$options" == "collapse_trunc" ]]; then cat {output.col_R} {output.col_trunc} > {output.R}; elif [[ "$options" == "all" ]]; then cat {output.col_R} {output.col_trunc} {output.R1} {output.R2} {output.strunc} > {output.R}; fi; """ |
213 214 215 216 217 218 219 | shell: """ out={output.R1}; AdapterRemoval --threads {threads} {params} --file1 {input[0]} \ --file2 {input[1]} --basename ${{out%%_R1.fastq.gz}} --gzip \ --output1 {output.R1} --output2 {output.R2} 2> {log}; """ |
254 255 256 257 258 259 260 261 262 263 | shell: """ ## remove --collapse from $params (needed for SE libs in a paired-end setting) params=$(echo {params} | sed 's/ --collapse[^ ]*//g') out={output.R}; AdapterRemoval --threads {threads} $params --file1 {input} \ --basename ${{out%%.fastq.gz}} --gzip \ --output1 {output.R} 2> {log}; """ |
297 298 299 300 301 302 | shell: """ set +e; AdapterRemoval --threads {threads} {params.params} --file1 {input[0]} \ --file2 {input[1]} --identify-adapters > {output}; """ |
338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 | shell: """ fastp --in1 {input[0]} --in2 {input[1]} --out1 {output.R1} --out2 {output.R2} \ --merged_out {output.R_merged} \ --json {output.json} --html {output.html} --thread {threads} {params} \ -R "Fastp report of {wildcards.sm}/{wildcards.lb}/{wildcards.id}" 2> {log}; ## what should be retained options={params.collapsed}; if [[ "$options" == "only_collapse" ]]; then ln -srf {output.R_merged} {output.R}; elif [[ "$options" == "all" ]]; then cat {output.R_merged} {output.R1} {output.R2} > {output.R}; fi; """ |
384 385 386 387 388 389 | shell: """ fastp --in1 {input[0]} --in2 {input[1]} --out1 {output.R1} --out2 {output.R2} \ --json {output.json} --html {output.html} --thread {threads} {params} \ -R "Fastp report of {wildcards.sm}/{wildcards.lb}/{wildcards.id}" 2> {log}; """ |
420 421 422 423 424 425 | shell: """ fastp --in1 {input} --out1 {output.R} \ --json {output.json} --html {output.html} --thread {threads} {params} \ -R "Fastp report of {wildcards.sm}/{wildcards.lb}/{wildcards.id}" 2> {log}; """ |
468 469 470 471 | shell: """ bwa aln {params} -t {threads} {input.ref} -f {output} {input.fastq} 2> {log} """ |
509 510 511 512 | shell: """ bwa aln {params} -t {threads} {input.ref} -f {output} {input.fastq} 2> {log} """ |
553 554 555 556 557 558 | shell: """ (bwa samse {params.params} \ -r \"@RG\\tID:{wildcards.id}\\tLB:{wildcards.lb}\\tSM:{wildcards.sm}\\tPL:{params.PL}\" \ {input.ref} {input.sai} {input.fastq} | samtools view -Sb > {output}) 2> {log} """ |
601 602 603 604 605 606 607 | shell: """ (bwa sampe {params.params} \ -r \"@RG\\tID:{wildcards.id}\\tLB:{wildcards.lb}\\tSM:{wildcards.sm}\\tPL:{params.PL}\" \ {input.ref} {input.sai1} {input.sai2} {input.fastq} | \ samtools view -Sb > {output}) 2> {log} """ |
647 648 649 650 651 652 | shell: """ bwa mem {params.params} -t {threads} \ -R \"@RG\\tID:{wildcards.id}\\tLB:{wildcards.lb}\\tSM:{wildcards.sm}\\tPL:{params.PL}\" \ {input.ref} {input.fastq} 2> {log} | samtools view -bS --threads {threads} - > {output}; """ |
690 691 | script: "../scripts/mapping_bowtie2.py" |
718 719 720 721 | shell: """ samtools sort --threads {threads} {input} > {output} 2> {log} """ |
760 761 762 763 764 | shell: """ samtools view -b --threads {threads} {params} \ -U {output.low_qual} {input} > {output.mapped} 2> {log} """ |
796 797 798 799 | shell: """ samtools view -b --threads {threads} {params} {input} > {output.mapped} 2> {log} """ |
86 87 88 89 90 91 92 93 94 95 96 | shell: """ ln -srf {input} {output.panel_vcf}; ## if index does not exist create it if [ -f "{input}.csi" ]; then ln -srf {input}.csi {output.panel_vcf_csi}; else bcftools index -f {output.panel_vcf} -o {output.panel_vcf_csi}; fi """ |
109 110 111 112 | shell: """ ln -srf {input} {output}; """ |
152 153 154 155 156 157 158 159 160 161 162 | shell: """ bcftools view -G -m 2 -M 2 -v snps \ {input.vcf} \ -Oz -o {output.sites}; bcftools index -f {output.sites}; bcftools query -f'%CHROM\\t%POS\\t%REF,%ALT\\n' {output.sites} | \ bgzip -c > {output.tsv}; tabix -s1 -b2 -e2 {output.tsv}; """ |
195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 | shell: """ header={output.final_vcf}.header; vcf_tmp={output.final_vcf}.tmp; bcftools mpileup -f {input.ref} \ -I -E -a 'FORMAT/DP' \ -T {input.sites} -r {wildcards.chr} \ -Ob --threads {threads} \ --ignore-RG {input.bam} | \ bcftools call -Aim -C alleles -T {input.tsv} -Oz -o ${{vcf_tmp}}; echo {input.bam} {wildcards.sm} > ${{header}}; bcftools reheader -s ${{header}} -o {output.final_vcf} ${{vcf_tmp}}; bcftools index -f {output.final_vcf}; rm -f ${{header}} ${{vcf_tmp}}; """ |
242 243 244 245 246 247 248 | shell: """ GLIMPSE_chunk {params.params} \ --input {input} \ --region {wildcards.chr} \ --output {output.chunks} """ |
289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 | shell: """ ## get chunk LINE=$( head -n {wildcards.n} {input.chunks} |tail -n1 ); ## run glimpse GLIMPSE_phase {params.params} \ --input {input.vcf_sample} \ --reference {input.vcf_ref} \ --map {input.map} \ --input-region $(echo $LINE | cut -d" " -f3) \ --output-region $(echo $LINE | cut -d" " -f4) \ --output {output.bcf} \ --thread {threads}; ## index bcf file bcftools index -f {output.bcf}; """ |
344 345 346 347 348 349 350 351 | shell: """ list_files={output.ligated_bcf}.list_files; for file in {input.bcf}; do echo $file; done > ${{list_files}}; GLIMPSE_ligate --input ${{list_files}} --output {output.ligated_bcf}; rm ${{list_files}}; bcftools index -f {output.ligated_bcf}; """ |
386 387 388 389 390 | shell: """ bcftools concat {input.bcf} -Oz -o {output.bcf}; bcftools index -f {output.bcf}; """ |
413 414 415 416 417 | shell: """ bcftools view -i'FORMAT/GP[0:*]>{wildcards.gp}' -Ob -o {output.bcf} {input.bcf}; bcftools index -f {output.bcf}; """ |
439 440 441 442 | shell: """ bcftools query -f '[%GT]\\t[%GP]\\n' {input.bcf} > {output.table}; """ |
477 478 479 480 481 482 483 484 485 486 487 | shell: """ Rscript {params.script} \ --input={input} \ --output={output} \ --width={params.width} \ --height={params.height} \ --gp={params.gp} \ --sample={wildcards.sm} \ --genome={wildcards.genome} """ |
506 507 508 509 510 511 512 | shell: """ GLIMPSE_sample --input {input.ligated_bcf} \ --solve \ --output {output.phased_bcf}; bcftools index -f {output.phased_bcf}; """ |
42 43 | script: "../scripts/fasta_indexing.py" |
80 81 | script: "../scripts/fasta_indexing.py" |
108 109 | script: "../scripts/fasta_indexing.py" |
137 138 | script: "../scripts/fasta_indexing.py" |
165 166 | shell: "samtools index {input} {output} 2> {log};" |
190 191 | shell: "samtools index {input} {output} 2> {log};" |
26 27 28 29 | shell: """ samtools merge -f --threads {threads} {output} {input} 2> {log}; """ |
54 55 56 57 | shell: """ samtools merge -f --threads {threads} {output} {input} 2> {log}; """ |
95 96 97 98 99 | shell: """ {params.PICARD} MarkDuplicates --INPUT {input} --OUTPUT {output.bam} --METRICS_FILE {output.stats} \ {params.params} --ASSUME_SORT_ORDER coordinate --VALIDATION_STRINGENCY LENIENT 2> {log}; """ |
132 133 134 135 136 137 | shell: """ bam={output.bam} dedup -i {input} $params -o $(dirname $bam); mv ${{bam%%.bam}}_rmdup.bam $bam """ |
171 172 173 174 175 | shell: """ mapDamage -i {input.bam} -r {input.ref} -d $(dirname {output.deamination}) \ {params.params} --merge-reference-sequences 2> {log}; """ |
202 203 204 205 206 | shell: """ mapDamage -i {input.bam} -r {input.ref} -d $(dirname {input.deamination}) \ {params.params} --merge-reference-sequences --rescale-only --rescale-out {output} 2>> {log}; """ |
231 232 233 234 | shell: """ bam trimBam {input} {output} {params} 2>> {log}; """ |
28 29 30 31 | shell: """ samtools merge -f --threads {threads} {output} {input} 2> {log}; """ |
54 55 56 57 | shell: """ samtools merge -f --threads {threads} {output} {input} 2> {log}; """ |
90 91 92 93 94 95 | shell: """ {params.GATK} -I {input.bam} -R {input.ref} -T RealignerTargetCreator -o {output.intervals} 2> {log}; \ {params.GATK} -I {input.bam} -T IndelRealigner -R {input.ref} -targetIntervals \ {output.intervals} -o {output.bam} 2>> {log}; """ |
119 120 121 122 | shell: """ samtools calmd --threads {threads} {input.bam} {input.ref} 2> {log} | samtools view -bS - > {output} """ |
138 139 140 141 142 | run: if is_external_sample(wildcards.sm, wildcards.genome): shell("ln -srf {input} {output}") else: shell("cp {input} {output}") |
158 159 160 161 | shell: """ cp {input} {output} """ |
7 8 9 10 | shell: """ echo snakemake version $(snakemake --version) > {output} || echo snakemake not available > {output} """ |
20 21 22 23 | shell: """ AdapterRemoval --version 2> >(sed 's/ver./version/g') > {output} || echo AdapterRemoval not available > {output} """ |
33 34 35 36 37 38 39 40 41 42 43 | shell: """ set +e; txt=$(bwa 2>&1 | grep Version | sed 's/Version:/bwa version/g') if [[ "$txt" == *"bwa"* ]]; then echo $txt > {output}; else echo bwa not available > {output}; fi exit 0; """ |
53 54 55 56 57 58 59 60 61 62 63 | shell: """ set +e; txt=$(bowtie2 2>&1 | grep "Bowtie 2 version") if [[ "$txt" == *"Bowtie"* ]]; then echo $txt > {output}; else echo bowtie2 not available > {output}; fi exit 0; """ |
73 74 75 76 | shell: """ fastqc --version | sed 's/v/version /g' > {output} || echo fastqc not available > {output} """ |
86 87 88 89 | shell: """ echo GenomeAnalysisTK version $(GenomeAnalysisTK --version) > {output} || echo GenomeAnalysisTK not available > {output} """ |
99 100 101 102 | shell: """ echo mapDamage version $(mapDamage --version) > {output} || echo mapDamage not available > {output} """ |
114 115 116 117 118 119 120 121 122 123 124 | shell: """ set +e; txt=$({params.PICARD} MarkDuplicates --version 2>&1 | sed 's/Version:/Picard MarkDuplicates version /g') if [[ "$txt" == *"Picard"* ]]; then echo $txt > {output}; else echo Picard MarkDuplicates not available > {output}; fi exit 0; """ |
134 135 136 137 | shell: """ samtools version | head -n1| sed 's/samtools/samtools version/g' > {output} || echo samtools not available > {output} """ |
147 148 149 150 | shell: """ bcftools --version |grep bcftools | sed 's/bcftools/bcftools version/g' > {output} || echo bcftools not available > {output} """ |
160 161 162 163 164 165 166 167 168 169 170 | shell: """ set +e; txt=$(bam 2>&1 | grep Version | sed 's/Version:/BamUtil version/g') if [[ "$txt" == *"BamUtil"* ]]; then echo $txt > {output}; else echo BamUtil not available > {output}; fi exit 0; """ |
180 181 182 183 | shell: """ bedtools --version | sed 's/v/version /g' > {output} || echo bedtools not available > {output} """ |
193 194 195 196 197 198 199 200 201 202 203 | shell: """ set +e; txt=$(seqtk 2>&1 | grep Version | sed 's/Version:/seqtk version/g') if [[ "$txt" == *"seqtk"* ]]; then echo $txt > {output}; else echo seqtk not available > {output}; fi exit 0; """ |
213 214 215 216 217 218 219 220 221 222 223 | shell: """ set +e; txt=$(dedup --version 2>&1 | head -n1 | sed 's/v/version /g') if [[ "$txt" == *"DeDup"* ]]; then echo $txt > {output}; else echo DeDup not available > {output}; fi exit 0; """ |
233 234 235 236 | shell: """ qualimap --help | grep QualiMap | sed 's/v./version /g' > {output} || echo qualimap not available > {output} """ |
246 247 248 249 | shell: """ multiqc --version | sed 's/,//g' > {output} || echo multiqc not available > {output} """ |
259 260 261 262 | shell: """ GLIMPSE_chunk --help | grep Version | sed 's/ \* Version :/glimpse version/g' > {output} || echo glimpse not available > {output} """ |
272 273 274 275 | shell: """ fastp --version 2> >(sed 's/fastp/fastp version/g') > {output} || echo fastp not available > {output} """ |
285 286 287 288 | shell: """ R --version | head -n 1 > {output} || echo R not available > {output} """ |
47 48 49 50 51 52 53 54 55 56 57 58 59 | shell: """ ## symlink file to remove '_R1' of paired reads html={output.html} symlink=${{html%%_fastqc.html}}.fastq.gz ln -srf {input.fastq[0]} $symlink ## run fastqc (-t 2 is a trick to increase the memory allocation / multitreathing is per file, so still only 1CPU used) fastqc --quiet -t 2 --outdir $(dirname $html) $symlink 2> {log}; ## remove symlink again rm $symlink """ |
85 86 | shell: "samtools flagstat --threads {threads} {input} > {output} 2> {log};" |
112 113 | shell: "samtools stats --threads {threads} {input} > {output} 2> {log};" |
136 137 138 139 | shell: """ bedtools genomecov -ibam {input} > {output} """ |
164 165 166 167 | shell: """ samtools view {input} | perl {params.script} -o {output} >> {log} """ |
191 192 193 194 | shell: """ samtools idxstats {input.bam} > {output} """ |
227 228 229 230 231 232 233 | shell: """ Rscript {params.script} \ --idxstats={input} \ --out={output} \ {params.sex_params} """ |
243 244 245 246 247 | shell: """ echo "Sex,Rx,CI,signif_set,reads_autosomes,reads_X" > {output} echo "NaN,NaN,NaN,NaN,NaN,NaN" >> {output} """ |
273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 | shell: """ Rscript {params.script} \ --id={wildcards.id} \ --lb={wildcards.lb} \ --sm={wildcards.sm} \ --genome={wildcards.genome} \ --output_file={output} \ --path_fastqc_orig={input.fastqc_orig} \ --path_fastqc_trim={input.fastqc_trim} \ --path_stats_mapped_highQ={input.stats_mapped_highQ} \ --path_length_mapped_highQ={input.length_fastq_mapped_highQ} \ --script_parse_fastqc={params.script_parse_fastqc} """ |
317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 | shell: """ list_fastq_stats=$(echo {input.fastq_stats} |sed 's/ /,/g'); if [ {params.chrs_selected} == "not_requested" ] then chrsSelected="" else chrsSelected="--chrs_selected={params.chrs_selected}" fi Rscript {params.script} \ --lb={wildcards.lb} \ --sm={wildcards.sm} \ --genome={wildcards.genome} \ --output_file={output} \ --path_list_stats_fastq=${{list_fastq_stats}} \ --path_stats_unique={input.stats_unique} \ --path_length_unique={input.length_unique} \ --path_idxstats_unique={input.idxstats_unique} \ $chrsSelected """ |
373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 | shell: """ list_lb_stats=$(echo {input.lb_stats} |sed 's/ /,/g'); if [ {params.chrs_selected} == "not_requested" ] then chrsSelected="" else chrsSelected="--chrs_selected={params.chrs_selected}" fi Rscript {params.script} \ --sm={wildcards.sm} \ --genome={wildcards.genome} \ --output_file={output} \ --path_list_stats_lb=$list_lb_stats \ --path_length_unique={input.length_unique} \ --path_idxstats_unique={input.idxstats_unique} \ --path_sex_unique={input.sex_unique} \ $chrsSelected """ |
410 411 412 413 414 415 | run: import pandas as pd df_list = [pd.read_csv(file) for file in input] df = pd.concat(df_list) df.to_csv(str(output), index=False) |
436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 | run: import pandas as pd import re def get_csv(file): my_csv = pd.read_csv(file) genome = re.sub(".*_stats.", "", file).replace(".csv", "") my_csv["genome"] = genome return my_csv df_list = [get_csv(file) for file in input] df = pd.concat(df_list) df.to_csv(str(output), index=False) |
476 477 478 479 480 481 482 | shell: """ Rscript {params.script} \ --path_genomecov={input} \ --sm={wildcards.sm} \ --output_file={output} """ |
496 497 498 499 500 501 502 503 | run: import pandas as pd df_list = [pd.read_csv(file) for file in input] # this should work even if the columns are not in the same order # across tables df = pd.concat(df_list) df.to_csv(str(output), index=False) |
554 555 556 557 558 559 560 561 562 563 564 565 566 567 568 569 570 571 572 573 | shell: """ ## get the subsampling interval nb=$(awk '{{sum += $3}} END {{print sum}}' {input.idxstats}); nth_line=1; # this part is to take a fraction of the total reads # to run bamdamage if [ {params.fraction} -eq 0 ]; then nth_line=1; elif [ {params.fraction} -lt 1 ]; then nth_line=$(( {params.fraction} * $nb )); elif [ {params.fraction} -lt "$nb" ]; then nth_line=$(( $nb / {params.fraction} )); fi; perl {params.script} {params.params} \ --nth_read $nth_line --output {output.damage_pdf} \ --output_length {output.length_pdf} {input.bam} 2>> {log}; """ |
606 607 608 609 610 611 612 613 614 615 616 617 618 619 620 621 622 623 624 625 | shell: """ ## extract the plot_length plot_length=$(echo {params.params} | sed 's/=/ /g' | awk -F '--plot_length' '{{print $2}}' | awk '{{print $1}}') if [ "$plot_length" != "" ]; then plot_length=--plot_length=$plot_length; fi Rscript {params.script} \ --length={input.length_table} \ --five_prime={input.dam_5prime_table} \ --three_prime={input.dam_3prime_table} \ --sample={wildcards.sm} \ --library={wildcards.lb} \ --genome={wildcards.genome} \ --length_svg={output.length} \ --damage_svg={output.damage} \ $plot_length ## delete the unwanted created Rplots.pdf... rm -f Rplots.pdf """ |
697 698 699 700 701 702 703 704 705 706 707 708 709 710 711 712 713 714 715 | shell: """ Rscript {params.script} \ --sm={input.sample_stats} \ --out_1_reads={output.plot_1_nb_reads} \ --out_2_mapped={output.plot_2_mapped} \ --out_3_endogenous={output.plot_3_endogenous} \ --out_4_duplication={output.plot_4_duplication} \ --out_5_AvgReadDepth={output.plot_5_AvgReadDepth} \ --out_6_Sex={output.plot_6_Sex} \ --x_axis={params.x_axis} \ --n_col={params.n_col} \ --color={params.color} \ --thresholds='{params.sex_thresholds}' \ --sex_ribbons='{params.sex_ribbons}' \ --width={params.width} \ --height={params.height} \ --show_numbers={params.show_numbers} """ |
742 743 744 745 | shell: """ qualimap bamqc -c -bam {input} -outdir {output} -nt {threads} --java-mem-size={resources.memory}M > {log} """ |
777 778 779 780 781 782 783 784 785 786 787 | shell: """ ## run mutliqc multiqc -c {params.config} \ -f \ -n $(basename {output.html}) \ -o $(dirname {output.html}) \ --title 'Mapache report (genome {wildcards.genome})' \ --cl-config "extra_fn_clean_trim: ['{params.resultdir}']" \ {input.files} 2> {log}; """ |
6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 | import subprocess import os ## get the original directory orig_fasta = snakemake.input.orig orig_prefix = os.path.splitext(orig_fasta)[0] ## get the extensions out_files = list(snakemake.output) file_extension = list(map(os.path.splitext, out_files)) ext = [el[1] for el in file_extension] ## check if the indexes are present files1 = [orig_fasta + s for s in ext] ## foo.fasta.ext files2 = [orig_prefix + s for s in ext] ## foo.ext if all([os.path.isfile(f) for f in files1]): ## foo.fasta.ext for idx, item in enumerate(files1): os.symlink(item, out_files[idx]) elif all([os.path.isfile(f) for f in files2]): ## foo.ext for idx, item in enumerate(files2): os.symlink(item, out_files[idx]) else: ## index is not present: create the index #print(eval(snakemake.params.cmd)) subprocess.run(eval(snakemake.params.cmd), shell=True) |
3 4 5 6 7 8 9 10 11 | import os if len(input.fastq) == 2: os.system(f'bowtie2 {snakemake.params.bowtie2_params} -p {snakemake.threads} -x {snakemake.input.ref} \ -1 {snakemake.input.fastq[0]} -2 {snakemake.input.fastq[1]} 2> {snakemake.log} | \ samtools view -bS - > {snakemake.output}') else: os.system(f'bowtie2 {snakemake.params.bowtie2_params} -p {snakemake.threads} -x {snakemake.input.ref} \ -U {snakemake.input.fastq} 2> {snakemake.log} | samtools view -bS - > {snakemake.output}') |
3 4 5 6 7 8 | import os if snakemake.params.run and snakemake.params.number != 0: os.system(f'seqtk sample {snakemake.params.params} {snakemake.input} {snakemake.params.number} | gzip > {snakemake.output}') else: os.system(f'ln -srf {snakemake.input} {snakemake.output}') |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 528 529 530 531 532 533 534 535 536 537 538 539 540 541 542 543 544 545 546 547 548 549 550 551 552 553 554 555 556 557 558 559 560 561 562 563 564 565 566 567 568 569 570 571 572 573 574 575 576 577 578 579 580 581 582 583 584 585 586 587 588 589 590 591 592 593 594 595 596 597 598 599 600 601 602 603 604 605 606 607 608 609 610 611 612 613 614 615 616 617 618 619 620 621 622 623 624 625 626 627 628 629 630 631 632 633 634 635 636 637 638 639 640 641 642 643 644 645 646 647 648 649 650 651 652 653 654 655 656 657 658 659 660 661 662 663 664 665 666 667 668 669 670 671 672 673 674 675 676 677 678 679 680 681 682 683 684 685 686 687 688 689 690 691 692 693 694 695 696 697 698 699 700 701 702 703 704 705 706 707 708 709 710 711 712 713 714 715 716 717 718 719 720 721 722 723 724 725 726 727 728 729 730 731 732 733 734 735 736 737 738 739 740 741 742 743 744 745 746 747 748 749 750 751 752 753 754 755 756 757 758 759 760 761 762 763 764 765 766 767 768 769 770 771 772 773 774 775 776 777 778 779 780 781 782 783 784 785 786 787 788 789 790 791 792 793 794 795 796 797 798 799 800 801 802 803 804 805 806 807 808 809 810 811 812 813 814 815 816 817 818 819 820 821 822 823 824 825 826 827 828 829 830 831 832 833 834 835 836 837 838 839 840 841 842 843 844 845 846 847 848 849 850 851 852 853 854 855 856 857 858 859 860 861 862 863 864 865 866 867 868 869 870 871 872 873 874 875 876 877 878 879 880 881 882 883 884 885 886 887 888 889 890 891 892 893 894 895 896 897 898 899 900 901 902 903 904 905 906 907 908 909 910 911 912 913 914 915 916 917 918 919 920 921 922 923 924 925 926 927 928 929 930 931 932 933 934 935 936 937 938 939 940 941 942 943 944 945 946 947 948 949 950 951 952 953 954 955 956 957 958 959 960 961 962 963 964 965 966 967 968 969 970 971 972 973 974 975 976 977 978 979 980 981 982 983 984 985 986 987 988 989 990 991 992 993 994 995 996 | from tkinter import E import pandas as pd import numpy as np import itertools import pathlib import re import subprocess, os.path from snakemake.io import Wildcards ########################################################################################## ## VERBOSE LOG ########################################################################################## ## print a summary of the contents def write_log(): ## reference genome if len(GENOMES) == 1: LOGGER.info(f"REFERENCE GENOME: 1 genome is specified:") elif len(GENOMES) < 4: LOGGER.info(f"REFERENCE GENOME: {len(GENOMES)} genomes are specified:") else: LOGGER.info(f"REFERENCE GENOME: {len(GENOMES)} genomes are specified.") ## get all chromosome names and store them in the dict config[chromosomes][genome][all] for later use for i, genome in enumerate(GENOMES): if i < 4: if len(get_param(["chromosome", genome], [])) > 1: LOGGER.info(f" - {genome}:") else: LOGGER.info(f" - {genome}") name = get_param(["chromosome", genome, "name"], "") if name: all_sex_chr = get_param(["chromosome", genome, "all_sex_chr"], "") LOGGER.info( f" - Detected genome as '{name}': Applying default chromosome names for sex and mt chromosomes" ) sex_chr = get_param(["chromosome", genome, "sex_chr"], "") if sex_chr: LOGGER.info(f" - Sex chromosome (X): {to_list(sex_chr)}") autosomes = get_param(["chromosome", genome, "autosomes"], "") if autosomes: if len(autosomes) > 10: LOGGER.info( f" - Autosomes: {autosomes[:4] + ['...'] + autosomes[-4:]}" ) else: LOGGER.info(f" - Autosomes: {autosomes}") elif i == 4: LOGGER.info(f" - ...") else: break # ---------------------------------------------------------------------------------------------------------- ## sample file LOGGER.info(f"SAMPLES:") if len(SAMPLES): if PAIRED_END: if SAMPLE_FILE == "yaml": LOGGER.info(f" - Samples (YAML input) in paired-end format:") else: LOGGER.info(f" - Sample file ('{SAMPLE_FILE}') in paired-end format:") LOGGER.info(f" - {len(SAMPLES)} SAMPLES") LOGGER.info(f" - {len([l for s in SAMPLES.values() for l in s])} libraries") tmp = [ i["Data2"] for s in SAMPLES.values() for l in s.values() for i in l.values() ] LOGGER.info( f" - {len([x for x in tmp if str(x) != 'nan'])} paired-end fastq files" ) nb = len([x for x in tmp if str(x) == "nan"]) if nb: LOGGER.info(f" - {nb} single-end fastq files") else: if SAMPLE_FILE == "yaml": LOGGER.info(f" - Samples (YAML input) in single-end format:") else: LOGGER.info(f" - Sample file ('{SAMPLE_FILE}') in single-end format:") LOGGER.info(f" - {len(SAMPLES)} SAMPLES") LOGGER.info(f" - {len([l for s in SAMPLES.values() for l in s])} libraries") LOGGER.info( f" - {len([i for s in SAMPLES.values() for l in s.values() for i in l])} single-end fastq files" ) if len(EXTERNAL_SAMPLES): if EXTERNAL_SAMPLE_FILE == "yaml": LOGGER.info(f" - External samples (YAML input; only stats are computed):") else: LOGGER.info( f" - External sample file ('{EXTERNAL_SAMPLE_FILE}'; only stats are computed):" ) for i, genome in enumerate(EXTERNAL_SAMPLES): if i < 4: LOGGER.info( f" - {len(EXTERNAL_SAMPLES[genome])} bam files {to_list(genome)}" ) elif i == 4: LOGGER.info(f" - ...") else: break # ---------------------------------------------------------------------------------------------------------- if len(final_bam): # print a summary of the workflow LOGGER.info("MAPPING WORKFLOW:") if run_subsampling: if type(subsampling_number) is str: LOGGER.info(f" - Subsampling with group specific settings") elif subsampling_number < 1: LOGGER.info(f" - Subsampling {100 * subsampling_number}% of the reads") else: LOGGER.info(f" - Subsampling {subsampling_number} reads per fastq file") run_adapter_removal = get_param(["adapterremoval", "run"], "True") ## get param 'simple' if type(run_adapter_removal) is dict: if COLLAPSE: LOGGER.info(f" - Removing adapters (variable) with AdapterRemoval and collapsing paired-end reads") else: LOGGER.info(f" - Removing adapters (variable) with AdapterRemoval") elif str2bool(run_adapter_removal): if COLLAPSE: LOGGER.info(f" - Removing adapters with AdapterRemoval and collapsing paired-end reads") else: LOGGER.info(f" - Removing adapters with AdapterRemoval") LOGGER.info(f" - Mapping with {mapper}") LOGGER.info(f" - Sorting bam file") if run_filtering: if save_low_qual: LOGGER.info( f" - Filtering and keeping separately low quality/unmapped reads" ) else: LOGGER.info(f" - Filtering and discarding low quality/unmapped reads") rmduplicates = get_param(["remove_duplicates", "run"], "markduplicates") ## get param 'simple' if rmduplicates == "markduplicates": LOGGER.info(f" - Removing duplicates with MarkDuplicates") elif rmduplicates == "dedup": LOGGER.info(f" - Removing duplicates with DeDup") if run_damage_rescale: LOGGER.info(f" - Rescaling damage with MapDamage2") if run_realign: LOGGER.info(f" - Realigning indels with GATK v3.8") if run_compute_md: LOGGER.info(f" - Recomputing md flag") # ---------------------------------------------------------------------------------------------------------- final_bam_tmp = final_bam + final_external_bam if len(final_bam_tmp): LOGGER.info("FINAL BAM FILES:") LOGGER.info( f" - BAM FILES ({len(final_bam_tmp)} files in folder '{os.path.dirname(final_bam_tmp[0])}'):" ) for i, file in enumerate(final_bam_tmp): if i < 4: LOGGER.info(f" - {file}") elif i == 4: LOGGER.info(f" - ...") else: break if save_low_qual: LOGGER.info( f" - LOW QUALITY AND UNMAPPED BAM FILES ({len(final_bam_low_qual)} files in folder '{os.path.dirname(final_bam_low_qual[0])}'):" ) for i, file in enumerate(final_bam_low_qual): if i < 4: LOGGER.info(f" - {file}") elif i == 4: LOGGER.info(f" - ...") else: break # ---------------------------------------------------------------------------------------------------------- LOGGER.info("ANALYSES:") if len(run_sex_inference): if len(genome): LOGGER.info(f" - Inferring sex {run_sex_inference}") else: LOGGER.info(f" - Inferring sex") if len(run_depth): if len(genome): LOGGER.info(f" - Computing depth per given chromosomes {run_depth}") else: LOGGER.info(f" - Computing depth per given chromosomes ") if run_damage != "False": if run_damage == "mapDamage": LOGGER.info( f" - Inferring damage and read length with MapDamage2 on all alignments" ) fraction = get_param(["damage", "bamdamage_fraction"], 0) if fraction == 0: LOGGER.info( f" - Inferring damage and read length with bamdamge on all alignments" ) elif fraction < 1: LOGGER.info( f" - Inferring damage and read length with bamdamge on {100 * fraction}% of the alignments" ) else: LOGGER.info( f" - Inferring damage and read length with bamdamge on {fraction} alignments" ) if len(run_imputation): if len(genome) > 1: LOGGER.info(f" - Imputing {run_imputation}") else: LOGGER.info(f" - Imputing") # ---------------------------------------------------------------------------------------------------------- # print a summary of the stats LOGGER.info("REPORTS:") LOGGER.info(f" - Mapping statistics tables:") for i, file in enumerate(stat_csv): LOGGER.info(f" - {file}") if run_qualimap: LOGGER.info( f" - Qualimap report: {len(qualimap_files)} report(s) on final bam files:" ) for i, file in enumerate(qualimap_files): if i < 4: LOGGER.info(f" - {file}") elif i == 4: LOGGER.info(f" - ...") else: break if run_multiqc: LOGGER.info( f" - Mulitqc report: {len(multiqc_files)} HTML report(s) combining statistics per genome:" ) for i, file in enumerate(multiqc_files): if i < 4: LOGGER.info(f" - {file}") elif i == 4: LOGGER.info(f" - ...") else: break LOGGER.info( f" => Snakemake report: run 'snakemake --report report.html' after finalization of the run to get the report" ) ########################################################################################################## ## helper function to deal with the SAMPLES nested dict ## get all values of the given key/column (multiple if it is at the lb or sm level) def get_sample_column(col, wc=None): if wc is None: grp = [id[col] for sm in SAMPLES.values() for lb in sm.values() for id in lb.values() if col in id] elif 'id' in wc.keys(): grp = [val for keys, val in SAMPLES[wc.sm][wc.lb][wc.id].items() if col in keys] elif 'lb' in wc.keys(): grp = [val[col] for val in SAMPLES[wc.sm][wc.lb].values() if col in val] elif 'sm' in wc.keys(): grp = [id[col] for lb in SAMPLES[wc.sm].values() for id in lb.values() if col in id] return grp ## get the number of fastq files of the given sample path def get_sample_count(wc=None): if wc is None: grpNb = len([id for sm in SAMPLES.values() for lb in sm.values() for id in lb.values()]) elif 'id' in wc.keys(): grpNb = 1 elif 'lb' in wc.keys(): grpNb = len([val for val in SAMPLES[wc.sm][wc.lb].values()]) elif 'sm' in wc.keys(): grpNb = len([id for lb in SAMPLES[wc.sm].values() for id in lb.values()]) return grpNb ## get the path, e.g., 'SAMPLES[sm][lb][id]' def get_sample_path(wc=None): if wc is None: return "SAMPLES" if 'id' in wc.keys(): return f"SAMPLES[{wc.sm}][{wc.lb}][{wc.id}]" if 'lb' in wc.keys(): return f"SAMPLES[{wc.sm}][{wc.lb}]" if 'sm' in wc.keys(): return f"SAMPLES[{wc.sm}]" ## return true if the data is paired-end and throw an error if it is a mixture def is_paired_end(wc): ## get all Data2 elements data2 = get_sample_column('Data2', wc) ## if not present it is SE if len(data2) == 0: return False ## if it is not homogenous across the group, return an error nbItems= get_sample_count(wc) if len(data2) != nbItems: LOGGER.error( f"ERROR: {get_sample_path(wc)} does not contain key 'Data2' in all rows ({len(data2)} of {nbItems} present)!" ) sys.exit(1) ## entry may be empty (SE): either all or none nbEmpty = data2.count('NULL') if nbEmpty == 0: return True if nbEmpty == nbItems: return False ## all are empty LOGGER.error( f"ERROR: {get_sample_path(wc)} has a mixture of single-end ({nbEmpty}) and paired-end ({nbItems-nbEmpty}) fastq files!" ) sys.exit(1) ## return true if the data is collapsed paired-end data and throw an error if it is a mixture def is_collapse(wc): ## do we have paired reads at the start? paired = is_paired_end(wc) if not paired: return False ## does the cleaning collapses the paired reads?: check if collapse is mentioned in the param run_cleaning = get_paramGrp(["cleaning", "run"], ["adapterremoval", "fastp", "False"], wc) if run_cleaning == "adapterremoval": params = get_paramGrp(["cleaning", "params_adapterremoval"], "--minlength 30 --trimns --trimqualities", wc) return any(ext in params for ext in ["--collapse", "--collapse-deterministic", "--collapse-conservatively"]) elif run_cleaning == "fastp": params = get_paramGrp(["cleaning", "params_fastp"], "--minlength 30 --trimns --trimqualities", wc) return any(ext in params for ext in ["--merge", "-m"]) else: return False ####################################################################################################################### ## read sample file def read_sample_file(): file = get_param(["sample_file"], "") #print(file) if file == "": return {}, "" if type(file) is dict: ## yaml input SAMPLES = file input = "yaml" else: ## sample file ## read the file delim = get_param(["delim"], "\s+") db = pd.read_csv(file, sep=delim, comment="#", dtype=str, keep_default_na=False) input = file ## check number of columns and column names colsSE = ["SM", "LB", "ID", "Data"] colsPE = ["SM", "LB", "ID", "Data1", "Data2"] if not set(colsSE).issubset(db.columns) and not set(colsPE).issubset(db.columns): LOGGER.error( f"ERROR: The column names in the sample file are wrong: single-end: {colsSE}; paired-end: {colsPE}" ) sys.exit(1) ## -------------------------------------------------------------------------------------------------- ## check if all IDs per LB and SM are unique all_fastq = db.groupby(["ID", "LB", "SM"])["ID"].agg(["count"]).reset_index() if max(all_fastq["count"]) > 1: LOGGER.error( f"ERROR: The ID's {all_fastq['ID']} are not uniq within library and sample!" ) sys.exit(1) ## -------------------------------------------------------------------------------------------------- ## dataframe to nested dict SAMPLES = {} cols = [e for e in list(db.columns) if e not in ("SM", "LB", "ID")] for index, row in db.iterrows(): ## if key not present add new dict if row["SM"] not in SAMPLES: SAMPLES[row["SM"]] = {} if row["LB"] not in SAMPLES[row["SM"]]: SAMPLES[row["SM"]][row["LB"]] = {} if row["ID"] not in SAMPLES[row["SM"]][row["LB"]]: SAMPLES[row["SM"]][row["LB"]][row["ID"]] = {} ## add all remaining columns to this dict for col in cols: SAMPLES[row["SM"]][row["LB"]][row["ID"]][col] = row[col] # ## from dataframe to nested dict # from collections import defaultdict # d = defaultdict(dict) # cols = [e for e in list(db.columns) if e not in ("SM", "LB", "ID")] # for row in db.itertuples(index=False): # for col in cols: # d[row.SM][row.LB][row.ID][col] = row[col] # SAMPLES = dict(d) return SAMPLES, input def test_SAMPLES(): ## do we have paired-end data or single-end data PAIRED_END = len(get_sample_column("Data1")) > 0 ## test all fastq files if PAIRED_END: ## forward reads #print(get_sample_column("Data1")) for fq in get_sample_column("Data1"): if not os.path.isfile(fq) and fq[:3] != 'ftp': LOGGER.error(f"ERROR: Fastq file '{fq}' does not exist!") sys.exit(1) ## reverse reads (may be NaN) for fq in get_sample_column("Data2"): if fq != 'NULL' and not os.path.isfile(fq) and fq[:3] != 'ftp': LOGGER.error(f"ERROR: Fastq file '{fq}' does not exist!") sys.exit(1) else: ## single end for fq in get_sample_column("Data"): if not os.path.isfile(fq) and fq[:3] != 'ftp': LOGGER.error(f"ERROR: Fastq file '{fq}' does not exist!") sys.exit(1) ## test if collapsing is correctly set COLLAPSE = "--collapse" in get_param( ["adapterremoval", "params"], "--minlength 30 --trimns --trimqualities") ## do we have Group settings? test if 'default' is absent col = 'Group' groups = get_sample_column(col) if len(groups) != 0: ## the word 'default' is reserved and may not be used as keyword grpList = [ii.split(',') for ii in groups] if [x.count('default') for x in grpList].count(0) != len(grpList): LOGGER.error( f"ERROR: Sample file column 'Group' contains the word 'default' which is not an allowed keyword!" ) sys.exit(1) return PAIRED_END, COLLAPSE ########################################################################################## ## read config[external_sample] def get_external_samples(): external_sample = get_param(["external_sample"], "") if external_sample == "": return {}, "" if isinstance(external_sample, dict): samples_stats = external_sample EXTERNAL_SAMPLE_FILE = "yaml" elif os.path.isfile(external_sample): delim = get_param(["delim"], "\s+") db_stats = pd.read_csv(external_sample, sep=delim, comment="#", dtype=str) EXTERNAL_SAMPLE_FILE = external_sample colnames = ["SM", "Bam", "Genome"] if not set(colnames).issubset(db_stats.columns): LOGGER.error( f"ERROR: The column names in the bam file (given by parameter config[external_sample]) are wrong! Expected are {colnames}!" ) sys.exit(1) ## from dataframe to nested dict from collections import defaultdict d = defaultdict(dict) for row in db_stats.itertuples(index=False): d[row.Genome][row.SM] = row.Bam samples_stats = dict(d) else: LOGGER.error( f"ERROR: The argument of parameter config[external_sample] is not valide '{external_sample}'!" ) sys.exit(1) ## test for each GENOMES separately for genome in list(samples_stats): ## does the GENOMES name exist? if genome not in GENOMES: LOGGER.error( f"ERROR: Genome name config[external_sample] does not exist ({genome})!" ) sys.exit(1) ## test if there are duplicated sample names if len(list(samples_stats[genome])) != len(set(list(samples_stats[genome]))): LOGGER.error( f"ERROR: Parameter config[external_sample][{genome}] contains duplicated sample names!" ) sys.exit(1) ## test each bam file for id,bam in list(samples_stats[genome].items()): if not os.path.isfile(bam): LOGGER.error( f"ERROR: Bam file config[external_sample][genome][{id}][bam] does not exist ({bam})!" ) sys.exit(1) return samples_stats, EXTERNAL_SAMPLE_FILE ########################################################################################## ## all functions for main snakemake file ## get the argument of the keys (recursively acorss teh keays) ## keys: list of keys ## def_value: ## - the default value if the parameter is not specified ## - if a list: possible arguments (throw error if different), first element is default value def get_param(keys, def_value, my_dict=config): assert type(keys) is list ## check if only a default value is passed or a list of possible values (first one is default arg) if type(def_value) is list and len(def_value) > 1: def_valueI = def_value[0] else: def_valueI = def_value ## run recursively if len(keys) == 1: arg = my_dict.get(keys[0], def_valueI) else: arg = get_param(keys[1:], def_valueI, my_dict=my_dict.get(keys[0], {})) arg = eval_param(arg) ## check if the arg is within the list of possible values if type(def_value) is list and len(def_value) > 1: if str(arg) not in def_value: LOGGER.error( f"ERROR: The parameter config[{']['.join(keys)}] has no valid argument (currently '{arg}'; available {def_value})!" ) sys.exit(1) return arg ## same as get_param(), but arguments may be a dict with group specific settings ## group names may be specified in the sample file and may be any word, ## except 'default', which allows to define a default argument def get_paramGrp(keys, def_value, wc, my_dict=config): if type(def_value) is list and len(def_value) > 1: arg = get_param(keys, def_value[0], my_dict) else: arg = get_param(keys, def_value, my_dict) ## if it is not a dict: return value and stop here if type(arg) is not dict: ## check if the arg is within the list of possible values if type(def_value) is list and len(def_value) > 1: if str(arg) not in def_value: LOGGER.error( f"ERROR: The parameter config[{']['.join(keys)}] has no valid argument (currently '{arg}'; available {def_value})!" ) sys.exit(1) param = arg else: ## get all 'Group' keywords (of the sample file of the given rank (sm, lb or id) col = 'Group' if 'id' in wc.keys(): grp = [val for keys, val in SAMPLES[wc.sm][wc.lb][wc.id].items() if col in keys] grpName = f"SAMPLES[{wc.sm}][{wc.lb}][{wc.id}]" elif 'lb' in wc.keys(): grp = [val[col] for val in SAMPLES[wc.sm][wc.lb].values() if col in val] grpName = f"SAMPLES[{wc.sm}][{wc.lb}]" elif 'sm' in wc.keys(): grp = [id[col] for lb in SAMPLES[wc.sm].values() for id in lb.values() if col in id] grpName = f"SAMPLES[{wc.sm}]" grpList = [ii.split(',') for ii in grp] if len(grpList) == 0: LOGGER.error( f"ERROR: The parameter config[{']['.join(keys)}] has group specific settings, but no keywords are available. Is the column '{col}' missing in the sample file!" ) sys.exit(1) ## go through all keywords and get the correct argument if type(def_value) is list and len(def_value) > 1: param = def_value[0] ## system default argument else: param = def_value ## system default argument for keyword in arg.keys(): ## if a default is defined take it, but continue searching if keyword == "default": param = arg[keyword] continue ## get the number of occurrences of the given group keyword argCount = [x.count(keyword) for x in grpList].count(0) ## if not present continue if argCount == len(grpList): continue ## if present in all rows, take it if argCount == 0: param = arg[keyword] break ## if not present in all rows throw an error LOGGER.error( f"ERROR: The parameter config[{']['.join(keys)}] has group specific settings, but the keyword '{keyword}' is not omnipresent in '{grpName}' (only {argCount} of {len(grpList)})!" ) sys.exit(1) ## check if the arg is within the list of possible values if type(def_value) is list and len(def_value) > 1: if str(param) not in def_value: LOGGER.error( f"ERROR: The parameter config[{']['.join(keys)}] has no valid argument (currently '{param}'; available {def_value})!" ) sys.exit(1) #print(f"{param} <= {keys} of {grpList} | {grpName}") #print(f"{param} <= {keys}") return param ## same as above, but a boolean is returned ## if it has a dict (group specific setting) a True is returned ## used at teh beginning to get a global view what is used def get_param_bool(key, def_value, my_dict=config): arg = get_param(key, def_value[0], my_dict) ## search without predefined list (could be a dict...) if type(arg) is dict: return True if str(arg) not in def_value: LOGGER.error( f"ERROR: The parameter config[{']['.join(key)}] has no valid argument (currently '{arg}'; available {def_value})!" ) sys.exit(1) return str2bool(arg) def get_sex_threshold_plotting(): thresholds = { genome: get_param( keys=["sex_inference", genome, "thresholds"], def_value='list( "XX"=c(0.8, 1), "XY"=c(0, 0.6), "consistent with XX but not XY"=c(0.6, 1), "consistent with XY but not XX"=c(0, 0.8) )', ) for genome in GENOMES } sex_thresholds = "list({pair})".format( pair=", ".join([f'"{genome}"={thresholds[genome]}' for genome in GENOMES]) ) sex_thresholds = sex_thresholds.replace("=", "?") return sex_thresholds ## update the argument in a nested dict (and create leaves if needed) def update_value(keys, value, my_dict=config): key = keys[0] if len(keys) == 1: new_dict = {} new_dict[key] = value return new_dict else: if key in my_dict: new_dict = update_value(keys[1:], value, my_dict[key]) my_dict[key].update(new_dict) return my_dict else: my_dict[key] = {} new_dict = update_value(keys[1:], value, my_dict[key]) my_dict[key].update(new_dict) return my_dict ########################################################################################## ## functions to evaluate python code if necessary ## eval single element def eval_elem(x): try: if type(x) is str: return eval(x, globals=None, locals=None) else: return x except: return x def eval_param(x): if type(x) is list: return list(map(eval_elem, x)) elif type(x) is str: return eval_elem(x) else: return x def to_str(x): if type(x) is list: #return [str(i) for i in x] return list(map(str, x)) elif type(x) is int: return str(x) elif type(x) is float: return str(x) else: return x def to_list(x): if type(x) is list: return x return list(to_str(x).split(" ")) def is_external_sample(sample, genome): return genome in EXTERNAL_SAMPLES and sample in list(EXTERNAL_SAMPLES[genome]) ## convert python list to R vector def list_to_r_vector(x): return 'c("' + '","'.join(x) + '")' ########################################################################################## ## get all chromosome names of the given reference genome def get_chromosome_names(genome): return to_str(get_param(["chromosome", genome, "all"], "")) ## set the chromosome names from fasta and store them in config def set_chromosome_names(genome): ## test if fasta is valid fasta = get_param(["genome", genome], "") if not os.path.isfile(fasta): LOGGER.error( f"ERROR: Reference genome config[{genome}] does not exist ({fasta})!" ) ## get all chromosome names from the reference genome if pathlib.Path(f"{fasta}.fai").exists(): allChr = list( map(str, pd.read_csv(f"{fasta}.fai", header=None, sep="\t")[0].tolist()) ) elif pathlib.Path( f"{RESULT_DIR}/00_reference/{genome}/{genome}.fasta.fai" ).exists(): fasta = f"{RESULT_DIR}/00_reference/{genome}/{genome}.fasta" allChr = list( map(str, pd.read_csv(f"{fasta}.fai", header=None, sep="\t")[0].tolist()) ) else: cmd = f"grep '^>' {fasta} | cut -c2- | awk '{{print $1}}'" allChr = list( map(str, subprocess.check_output(cmd, shell=True, text=True).split()) ) config = update_value(["chromosome", genome, "all"], allChr) ## return a list of the chromosome names which do not match ## empty list means that all matched def valid_chromosome_names(genome, names): allChr = get_chromosome_names(genome) if type(names) is not list and names not in allChr: return [names] if list(set(names) - set(allChr)): return list(set(names) - set(allChr)) return [] ## check the chromosome names if they are valid (sex inference for this genome is set to true) def set_sex_inference(genome): ## get the chromosome names of the given genome allChr = get_chromosome_names(genome) ## get the specified sex and autosome chromosome names sex_chr = to_str(get_param(["sex_inference", genome, "sex_chr"], '')) autosomes = to_str(get_param(["sex_inference", genome, "autosomes"],[])) ## if the sex and autosome chromosome names are set, check if they make sense if sex_chr != '' and len(autosomes) > 0: # check if the chromosomes specified in sex determination exist ## X chromosome if len(sex_chr): #print(sex_chr) if valid_chromosome_names(genome, sex_chr): LOGGER.error( f"ERROR: Sex chromosome specified in config[sex_inference][{genome}][sex_chr] ({sex_chr}) does not exist in the reference genome." ) os._exit(1) config = update_value(["chromosome", genome, "sex_chr"], sex_chr) else: LOGGER.error( f"ERROR: No sex chromosome specified in config[sex_inference][{genome}][sex_chr]!" ) os._exit(1) # autosomes if len(autosomes): if valid_chromosome_names(genome, autosomes): LOGGER.error( f"ERROR: In config[sex_inference][{genome}][autosomes], the following chromosome names are not recognized: {valid_chromosome_names(genome, autosomes)}!" ) os._exit(1) config = update_value(["chromosome", genome, "autosomes"], autosomes) else: LOGGER.error( f"ERROR: No autosomes specified in config[sex_inference][{genome}][autosomes]!" ) os._exit(1) else: ## if they are not set, try to infer the genome (hg19 or GRCh38) hg19 = list(map(str, list(range(1, 23)) + ["X", "Y", "MT"])) GRCh38 = [f"chr{x}" for x in list(range(1, 23)) + ["X", "Y", "M"]] if set(hg19).issubset(set(allChr)): name = "hg19" detectedSexChrom = ["X", "Y", "MT"] detectedAutosomes = list(set(hg19) - set(detectedSexChrom)) elif set(GRCh38).issubset(set(allChr)): name = "GRCh38" detectedSexChrom = ["chrX", "chrY", "chrM"] detectedAutosomes = list(set(GRCh38) - set(detectedSexChrom)) else: LOGGER.error( f"ERROR: For sex inference the parameters config[sex_inference][{genome}][sex_chr] and config[sex_inference][{genome}][autosomes] are required!" ) os._exit(1) ## write the sex and autosome names config = update_value(["chromosome", genome, "name"], name) config = update_value(["chromosome", genome, "all_sex_chr"], detectedSexChrom) config = update_value(["chromosome", genome, "sex_chr"], detectedSexChrom[0]) config = update_value(["chromosome", genome, "autosomes"], detectedAutosomes) ## check if the specified chromosomes to compute the depth on are available def read_depth(genome): # check if chromosomes for which DoC was requested exist depth = to_str(get_param(["depth", genome, "chromosomes"], "")) if valid_chromosome_names(genome, depth): LOGGER.error( f"ERROR: config[depth][{genome}][chromosomes] contains unrecognized chromosome names ({valid_chromosome_names(genome, depth)})!" ) os._exit(1) config = update_value(["chromosome", genome, "depth"], depth) ## convert string to boolean def str2bool(v): return str(v).lower() in ("yes", "true", "t", "1") ## convert any argument to a list of string(s) def str2list(v): if type(v) is list: return [str(x) for x in v] else: return [str(v)] ## get incremental memory allocation when jobs fail ## 'startStr' is the first memory allocation in GB ## input is in GB; output in MB; default is 2GB, but can be changed by a rule ## get incremental memory allocation when jobs fail ## 'start' is the first memory allocation in GB (default 4GB) ## input is in GB; output is in MB; ## global variable memory_increment_ratio defines by how much (ratio) the memory is increased if not defined specifically def get_memory_alloc(module, attempt, default=2): moduleList = module if type(moduleList) is not list: moduleList = [module] mem_start = int(get_param(moduleList + ["mem"], default)) mem_incre = int( get_param( moduleList + ["mem_increment"], memory_increment_ratio * mem_start ) ) return int(1024 * ((attempt - 1) * mem_incre + mem_start)) ## in this second version the 'mem' is added to the word of the last element def get_memory_alloc2(module, attempt, default=2): moduleList = module if type(moduleList) is not list: moduleList = [moduleList] mem_start = int(get_param(moduleList[:-1] + [moduleList[-1] + "_mem"], default)) mem_incre = int( get_param( moduleList[:-1] + [moduleList[-1] + "_mem_increment"], memory_increment_ratio * mem_start, ) ) return int(1024 * ((attempt - 1) * mem_incre + mem_start)) def convert_time(seconds): day = seconds // (24 * 3600) seconds = seconds % (24 * 3600) hour = seconds // 3600 seconds %= 3600 minutes = seconds // 60 seconds %= 60 return "%d-%02d:%02d:%02d" % (day, hour, minutes, seconds) ## get incremental time allocation when jobs fail ## 'start' is the first time allocation in hours (default 12h) ## input is in hours; output is in minutes; ## global variable runtime_increment_ratio defines by how much (ratio) the time is increased if not defined specifically def get_runtime_alloc(module, attempt, default=12): moduleList = module if type(moduleList) is not list: moduleList = [module] time_start = int(get_param(moduleList + ["time"], default)) time_incre = int( get_param( moduleList + ["time_increment"], runtime_increment_ratio * time_start ) ) return int(60 * ((attempt - 1) * time_incre + time_start)) ## in this second version the 'time' is added to the word of the last element def get_runtime_alloc2(module, attempt, default=12): moduleList = module if type(moduleList) is not list: moduleList = [module] time_start = int( get_param(moduleList[:-1] + [moduleList[-1] + "_time"], default) ) time_incre = int( get_param( moduleList[:-1] + [moduleList[-1] + "_time_increment"], runtime_increment_ratio * time_start, ) ) return int(60 * ((attempt - 1) * time_incre + time_start)) ## get the number of threads of the given parameter def get_threads(module, default=1): moduleList = module if type(moduleList) is not list: moduleList = [module] return int(get_param(moduleList + ["threads"], default)) ## in this second version the 'threads' is added to the word of the last element def get_threads2(module, default=1): moduleList = module if type(moduleList) is not list: moduleList = [module] return int(get_param(moduleList[:-1] + [moduleList[-1] + "_threads"], default)) def bam2bai(bam): # return bam.replace('.bam', '.bai') return f"{bam[:len(bam) - 4]}.bai" ########################################################################################## ## check if java is called by a .jar file or by a wrapper def get_picard_bin(): bin = get_param(["software", "picard_jar"], "picard.jar") if bin[-4:] == ".jar": bin = "java -XX:ParallelGCThreads={threads} -XX:+UseParallelGC -XX:-UsePerfData \ -Xms{resources.memory}m -Xmx{resources.memory}m -jar bin" return bin def get_gatk_bin(): bin = get_param(["software", "gatk3_jar"], "GenomeAnalysisTK.jar") if bin[-4:] == ".jar": bin = "java -XX:ParallelGCThreads={threads} -XX:+UseParallelGC -XX:-UsePerfData \ -Xms{resources.memory}m -Xmx{resources.memory}m -jar bin" return bin |
1 of
scripts/utils.py
392 393 394 395 | shell: """ cat {input} > {output} """ |
412 413 414 415 416 | shell: """ for i in {input}; do echo $(grep 'Consensus:' $i | head -n1) {sm}/{lb}/{id}; done > {output.adapter1}; for i in {input}; do echo $(grep 'Consensus:' $i | tail -n1) {sm}/{lb}/{id}; done > {output.adapter2} """ |
Support
- Future updates
Related Workflows















